In ANPDB, each natural product is linked to its producer species and relevant references. You can begin a search by selecting the main icons located below the homepage or by navigating to the Search section in the taskbar.

If you know the name of the compound of interest, click on the Compounds icon on the homepage or Home -> Search -> Keyword, then just type in the name or synonym.

Furthermore, there is the possibility to search for compounds ordered alphabetically. To do so, navigate to Home -> Browse -> Compounds.

Sometimes, the name of a compound is not clearly defined or not known, but the structure of the compound is known. In that case it is possible to draw the structure by using the ChemDoodle structure editor. To do so, navigate to Home -> Search -> Similarity/Structure. It is not necessary to match the exact structure, similar structures will also be identified.

It is possible to change the search parameters e.g. decreasing the tanimoto coefficient in order to increase the number of retrieved similar compounds. Additionally, you can decide to search for substructures.

The second search option provides the possibility to search for the names of the source species alphabetically. Click on the Species icon on the homepage or Home -> Browse -> Species.

Click on the species name to be redirected to its detailed information page.

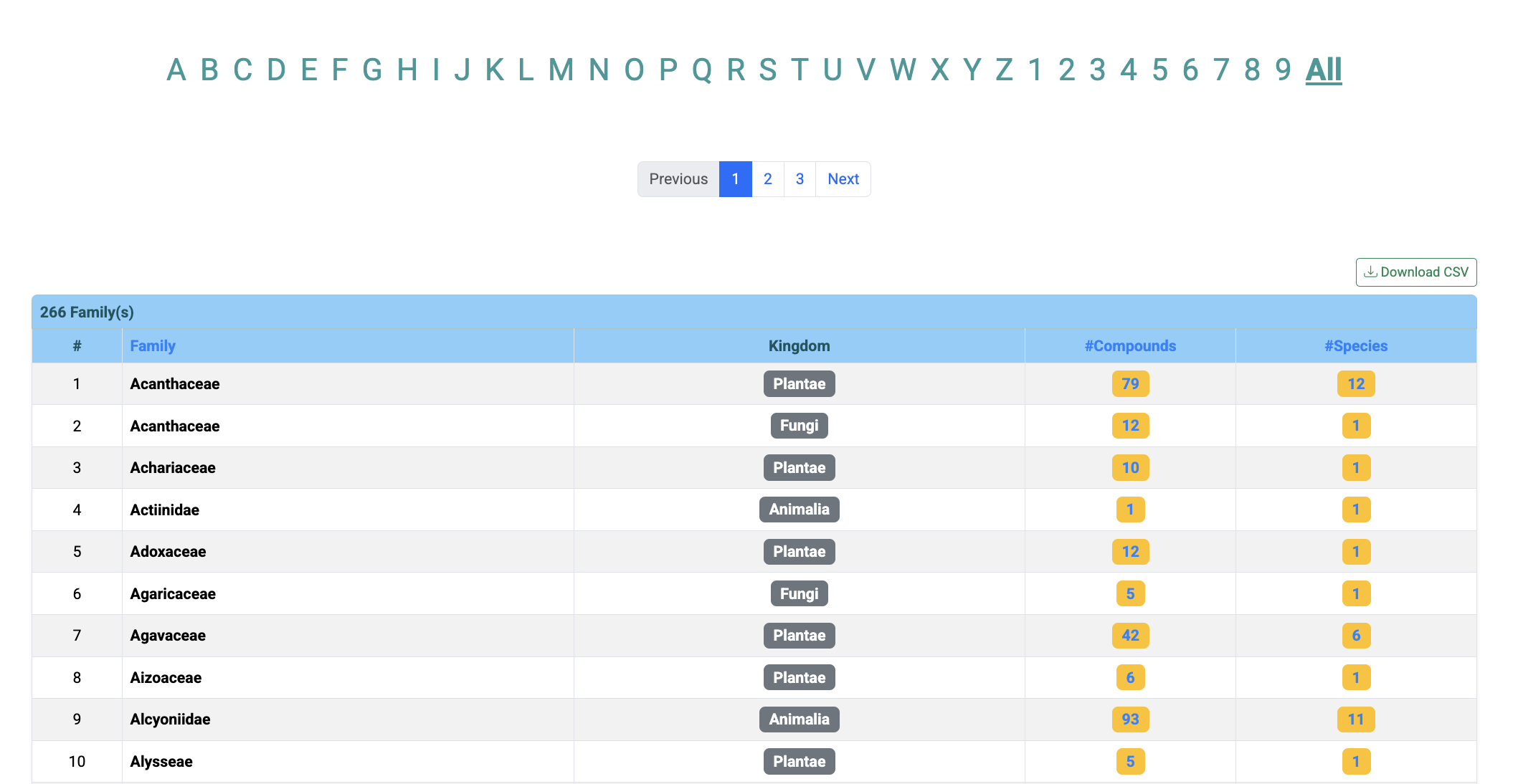
Compounds and species are also classified according to the family of the species. Click on Home -> Browse -> Families.
It is possible to search by references. Click on the Articles and Theses icon on the homepage or Home -> Browse -> References. PubMed IDs and Links of the describing articles are also available. Click on a PubMed ID or Link to access the article on the website.

ANPDB provides thousands of predicted NMR and MS spectra. Home -> Search -> Compounds (NMR/MS Data). You can specify multiplicity, chemical shift, and whether you are looking for 1H or 13C results for any number of signals. The multiplicity can be specified to a certain point (S = singlet; D = doublet; T = triplet; D.T = doublet of a triplet; T.T = triplet of a triplet), any other multiplicity can be found by choosing M (multiplet). You can also search for any number of MS peaks by mass-to-charge ratio (tolerance is 0.2 m/z).

The phylogenetic tree is based on species names and NCBI Taxon IDs. Home -> Browse -> Phylogeny. Clicking on the name "Halobacterium salinarum|2242" leads to the detailed information page of Halobacterium salinarum.

Similarity searches in ANPDB are conducted with Openbabel using 2D Daylight-like fingerprints or substructure searches. Those fingerprints represent structural features of a molecule. Fingerprints are dichotomous [0,1] (bit) arrays. Rather than encoding concrete structural features like structural keys do, there is no assigned meaning to each bit. Fingerprints are generated from the molecule itself using hash algorithms. There are
Subsequently, those patterns are generally encoded in four to five bits and added to the fingerprint. If patterns are substructures of another structure, all bits set in the substructure pattern's fingerprint will be set in the other's fingerprint. Boolean operations on those fingerprints provide an effective way for similarity calculations and substructure searches. Stereochemistry as well as atoms not connected to the "main" molecule are not considered in the kind of search ANPDB perfoms.
ANPDB uses the tanimoto coefficent for similarity calculations:
tcA,B=NA,B/(NA+NB-NA,B), with
You can search scaffolds by navigating to Home -> Search -> Scaffold. The search table displays all available scaffolds, starting from scaffold level 0. After selecting one or more structures, you can choose to either:

Several new features have been added to ANPDB. There are described in the section below.
The target prediction for compounds is performed using ePharmaLib. By clicking on the Predicted Protein Target on the compound card, the predicted binding targets are displayed in a table, sorted in descending order of binding probability based on the Tversky Score.

Click on Compound-Protein Relationships on the compound card to be redirected to the corresponding page in CPRiL.

On the first page, all functionally related proteins are listed. Click on the #Article(s) you are interested in to proceed to the second page.

On the second page, you will find all literatures documenting the relationship between this protein and the compound. Click on #Total Relationships to be redirected to the third page.

On the third page, you will find the exact sentences from this paper that describe the relationship between the protein and the compound.

NMR spectra were predicted with the cxcalc command-line tool and the interactive plots were generated with JSpecView. The user can switch between 1H, 13C or stacked 1H/13C NMR plots. Individual peak lists can be displayed after clicking on a given button below the spectrum.

Below is a display of the 1H NMR peak list with corresponding integration values (i.e., number of protons per signal).

MS fragmentation patterns were predicted using CMF-ID, and the interactive plots were generated with plotly.js and displayed in stacked format for 10V, 20V, and 40V. Hovering over a peak will display the predicted fragment structure along with the corresponding m/z value.

If you click on a compound name, ANPDB provides information about the compound, such as synonyms, physicochemical properties, related species, corresponding references, additional information about compound activity, scaffolds, NMR and MS spectra, Compound–Protein Relationships and Predicted Protein Targets.

ANPDB has been tested with IE8, FF3.5+, and webkit browsers (Safari, Firefox, Google Chrome). Although, all modern browsers of any brand should work fine.
Please cite:
African Natural Products Database (ANPDB): A Resource for Exploring the Therapeutic Potential of Natural Products from Africa
Jude Y. Betow, Ammar Qaseem, Boris D. Bekono, Kiran K. Telukunta, Aurélien F. A. Moumbock, Conrad V. Simoben, Smith B. Babiaka, Solange A. Tanyi, Vanessa A. Shu, Arianne T. Ndi, Pascal Amoa Onguéné, Clovis S. Metuge, Simeon Akame, Donatus B. Eni, Cyril T. Namba-Nzanguim, Mathieu J.M. Tjegbe, Marianka N. Dikande, Patience T. Akeh, Sounders Jr. Balgah, Cyprian D. Meh, Akachukwu Ibezim, Idris F. Tabi, Yvette I. Malange, Bakoh Ndingkokhar, Leonel E. Njume, Yue Feng, Said Amrani, Oyere T. Ebob, Wolfgang Sippl, Stefan Günther, and Fidele Ntie-Kang.
This manuscript is currently under revision for submission to NAR.
Pharmacoinformatic investigation of medicinal plants from East Africa.
Conrad V. Simoben, Ammar Qaseem, Aurélien F. A. Moumbock, Kiran K. Telukunta, Stefan Günther, Wolfgang Sippl and Fidele Ntie-Kang Molecular Informatics, 2020.
DOI: 10.1021/acs.jnatprod.7b00283
NANPDB: A Resource for Natural Products from Northern African Sources.
Fidele Ntie-Kang, Kiran K. Telukunta, Kersten Döring, Conrad V. Simoben, Aurélien F. A. Moumbock, Yvette I. Malange, Leonel E. Njume, Joseph N. Yong, Wolfgang Sippl, and Stefan GüntherJournal of Natural Products, 2017.
DOI: 10.1021/acs.jnatprod.7b00283